

Description:
In order to compute the flow around a molecule such as hemoglobin, it is necessary first to generate a surface triangulation of the molecule. The traditional approach has been to compute the intersection of surface triangulations for each sphere representing individual atoms in the molecule. The calculation of intersecting triangulations is a very complex problem and yields surface triangulations of very poor quality. Instead, we have developed a surface merging algorithm based on an adaptive voxelization followed by an iso-surface extraction. First a voxelizarion is created which covers the entire model. The the shortest distance of each voxel to the surface of each sphere is computed. The distance is assigned a negative sign if the voxel lies inside an atom or positive if it is outside. Once the distance map is calculated, the molecule surface is constructed by extracting the zero-distance iso-surface.
We start by constructing the surface triangulation for a single sphere (atom) and then combining several atoms together. The follogin two images show the surface triangulation of one sphere and the superposition of the first 20 atoms in the molecule.


The following series of images show the results obtained for these 20 spheres as the resolution of the voxelization is increased.
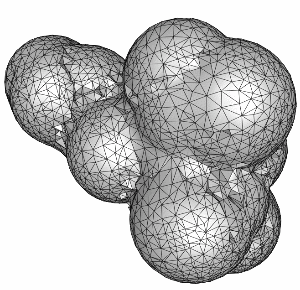
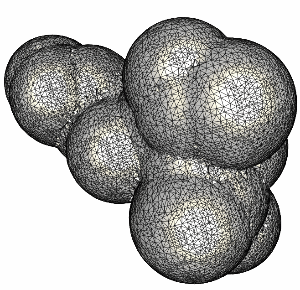
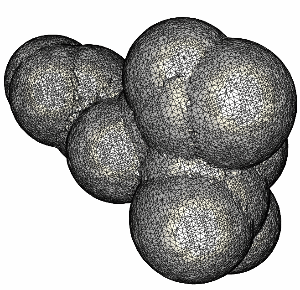

Finally, the surface of the complete molecule, consisting of 300 atoms is shown in the following figure.

Observations: